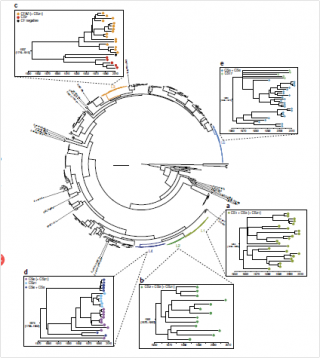
Enterotoxigenic E. coli (ETEC) bacteria cause annually at least 400 million diarrheal disease cases according to WHO investigations. The ETEC bacteria are a common trouble for travelers and kill approximately 400000 small children per year in the developing countries. According to current knowledge, these bacteria evolve very rapidly such that harmless commensals turn frequently into new ETEC strains via horizontal gene transfer. This belief was challenged in a study published in Nature Genetics, where a collection of global data covering 30 years and the whole genomes of hundreds of ETEC strains unearthed several stable evolutionary lineages in which the toxicity and virulence factors had remained virtually unchanged over several decades. Advanced computational modeling methods developed by the BSG group of professor Jukka Corander played an important role for this discovery. The study has considerable clinical implications, because it highlights that a majority of the ETEC disease cases would be preventable with a vaccine targeting the stable colonization factors of the major lineages identified from the high-density genome data. Given the anticipated success of such vaccines, more intensive resources for their development are politically motivated. For details, see the full article: Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nature Genetics. http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.3145.html
Last updated on 9 Dec 2014 by Jukka Corander - Page created on 9 Dec 2014 by Jukka Corander